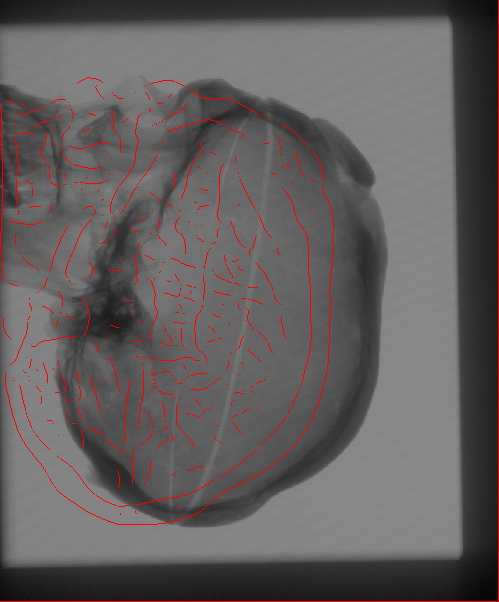
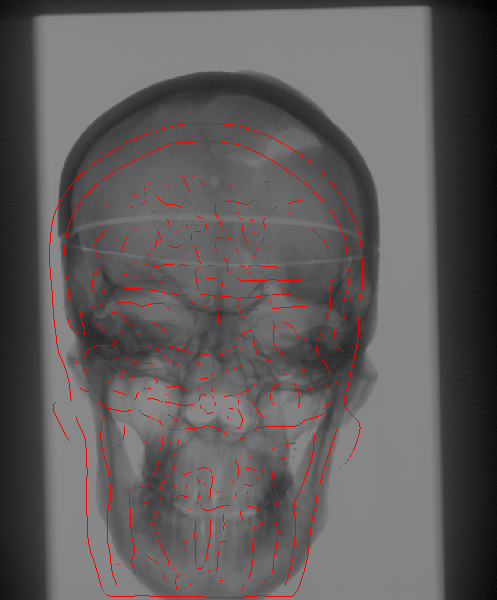
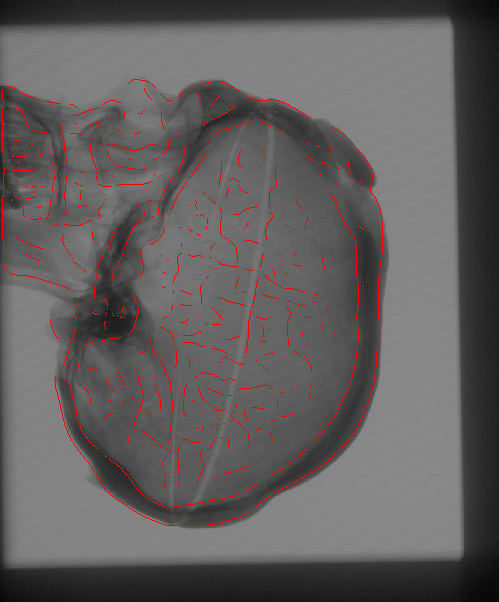
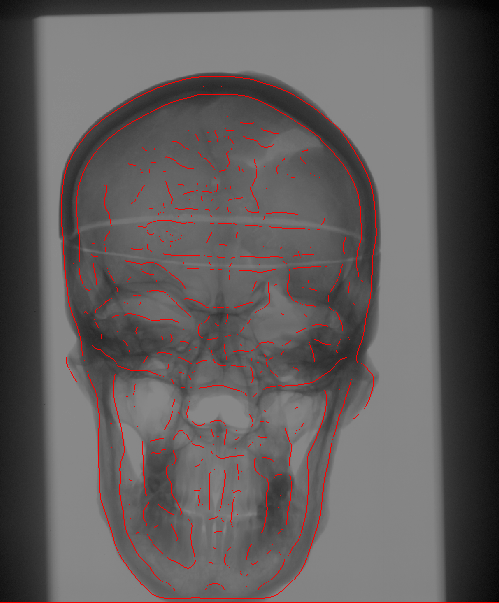
The Problem
Registration of two datasets is the process of identifying a
geometrical transformation that locates the coordinate system of one
in that of the other. Rigid body registration restricts the
searched transformation to be a combination of translations and
rotations which are sufficient to describe the movement of solid
objects.
We propose to register CT
volumetric datasets to corresponding 2D X-ray fluoroscopic
images. As a single 2D image, in practice, does not convey sufficient
information about the spatial location of the imaged object, we
require two projection images to achieve our task. We assume that the
two imaging views are related by a known transformation.
The main challenges of the problem lie in identifying a similarity
measure that can quantify the quality of the alignment between the
images and in defining a procedure to modify and refine current estimates
of the transformation parameters in a way that the similarity score is
optimized.
Motivation
Although 2D images lack significant information that is
present in 3D modalities, they might be conveniently and efficiently
used to record details about the most current state of the imaged
object. The most amount of information can be gained about the changes
recorded by the 2D modalities and the detailed 3D model if we fuse the
information provided by both of them. In order to achieve the proper
spatial alignment of the different components, it is necessary to
determine their relative position and orientation.
Approach
We propose an intensity-based registration algorithm using an
information theoretic objective function, mutual information (MI), to
establish the proper alignment of the input datasets. For
optimization purposes, we compare the performance of the non-gradient
Powell method and two slightly different versions of a stochastic
gradient ascent strategy: one using a sparsely sampled histogramming
approach and the other Parzen windowing to carry out probability
density approximation.
In order to compare the multi-dimensional images, with the current
estimate of the transformation parameters we create a simulated
version of the 2D acquisitions. A pair of such Digitally Reconstructed
Radiographs (DRRs) is compared to the observed X-ray images.
Our main contribution lies in adopting a stochastic approximation
scheme successfully applied in 3D-3D registration problems to the
2D-3D scenario, which obviates the need for the generation of full
DRRs at each iteration of pose optimization. This facilitates a
considerable savings in computation expense. We also introduce a new
probability density estimator for image intensities via sparse
histogramming, derive gradient estimates for the density measures
required by the maximization procedure and introduce the framework for
a multiresolution strategy to the problem.
The following link leads to animated gif files demonstrating the
incremental image aligning procedure.
(animation)




Lilla Zöllei,
Eric Grimson,
Alexander Norbash,
William Wells:
2D-3D Rigid Registration of X-Ray Fluoroscopy and CT Images Using Mutual Information
and Sparsely Sampled Histogram Estimators.
IEEE CVPR, 2001, to appear.
[PDF]
[Postscript]
Alexander Norbash anorbash@partners.org
Lilla Zöllei lzollei@ai.mit.edu
Back to the MIT AI Lab page.