 The Computer Vision Group of the MIT Artificial
Intelligence Lab has been collaborating closely for several years
with the Surgical
Planning Laboratory of Brigham and Women's Hospital. As part of
the collaboration, tools are being developed to support image guided
surgery. Such tools will enable surgeons to visualize internal
structures through an automated overlay of 3D reconstructions of
internal anatomy on top of live video views of a patient.
We are developing image analysis tools for leveraging the detailed
three-dimensional structure and relationships in medical images.
Sample applications are in preoperative surgical planning,
intraoperative surgical guidance, navigation, and instrument tracking.
The Computer Vision Group of the MIT Artificial
Intelligence Lab has been collaborating closely for several years
with the Surgical
Planning Laboratory of Brigham and Women's Hospital. As part of
the collaboration, tools are being developed to support image guided
surgery. Such tools will enable surgeons to visualize internal
structures through an automated overlay of 3D reconstructions of
internal anatomy on top of live video views of a patient.
We are developing image analysis tools for leveraging the detailed
three-dimensional structure and relationships in medical images.
Sample applications are in preoperative surgical planning,
intraoperative surgical guidance, navigation, and instrument tracking.

 The anatomical structures that appear in an internal scan such as MR
and CT must be explicitly extracted or segmented from the scan before
they can be directly used for surface registration or for 3D
visualization. By segmentation, we refer to the process of labeling
individual voxels in the volumetric scan by tissue type, based on
properties of the observed intensities as well as known anatomical
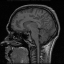
information about normal subjects. Below is an image of the raw MR
scan and an mpeg movie of all the sagittal slices.
The anatomical structures that appear in an internal scan such as MR
and CT must be explicitly extracted or segmented from the scan before
they can be directly used for surface registration or for 3D
visualization. By segmentation, we refer to the process of labeling
individual voxels in the volumetric scan by tissue type, based on
properties of the observed intensities as well as known anatomical
information about normal subjects. Below is an image of the raw MR
scan and an mpeg movie of all the sagittal slices.
The segmentation is performed using automated techniques and semi-automated techniques. Automatic segmentation techniques include an automatic gain artifact suppression technique based on expectation-maximization in association with a cortical volume isolation technique based on image morphology and active contours. Below is a slice of an MRI, with the brain, ventricles, and tumor segmented.
We use surface rendering techniques to display the segmented MRI structures. This procedure consists of first extracting bounding surfaces from the segmented MRI volume using the marching cubes algorithm. This algorithm generates a set of connected triangles to represent the 3D surface for each segmented structure. These surfaces are then displayed by selecting a virtual viewing camera location and orientation in the MRI coordinate frame and using standard computer graphics techniques to project the surface onto the viewing camera. This rendering process removes hidden portions of the surface, shades the surface according to its local normal, and optionally varies the surface opacity to allow glimpses into internal structures. Sample renderings and two movies are shown below.


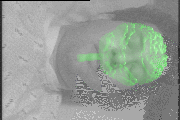
 After the registration, we have the transformation from the MRI
coordinate frame to the operating room coordinate frame--that is, we
know exactly where the MRI points are positioned in the patient--both
on the surface and internally. In the image below, we have blended
the 3D skin model with the video image of the patient. The movies
show the skin model being blended in and out to confirm the
registration.
After the registration, we have the transformation from the MRI
coordinate frame to the operating room coordinate frame--that is, we
know exactly where the MRI points are positioned in the patient--both
on the surface and internally. In the image below, we have blended
the 3D skin model with the video image of the patient. The movies
show the skin model being blended in and out to confirm the
registration.
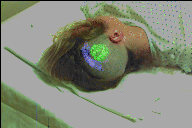
The registration points are overlaid on the 3D skin model as another method to verify the registration. The points are color coded based on the distance to the skin model (green = 0 mm, yellow = 2.5 mm, red = 5 mm).
![]() In addition to our intraoperative pointer, we have attached a bipolar
simulator (Cadwell Laboratories Inc., Washington, USA) to the
trackable probe (see image, right). This stimulator is used to
determine the location of vital regions of the brain, including motor
and sensory corticies and language area. When the stimulator is
placed on motor cortex, a muscle response occurs, and when placed on
sensory cortex, sensation in different areas is reported. Language
suppression (including temporary loss of speech) occurs when the
stimulator touches the languages area. As the neurosurgeon stimulates
different areas of the brain and receives responses, it is common for
him to place numbered markers on the cortex highlighting regions to
avoid. When our probe is attached to the stimulator, we can obtain
the position of the tip during stimulations and immediately produce a
color-coded visualization highlighting these important areas.
In addition to our intraoperative pointer, we have attached a bipolar
simulator (Cadwell Laboratories Inc., Washington, USA) to the
trackable probe (see image, right). This stimulator is used to
determine the location of vital regions of the brain, including motor
and sensory corticies and language area. When the stimulator is
placed on motor cortex, a muscle response occurs, and when placed on
sensory cortex, sensation in different areas is reported. Language
suppression (including temporary loss of speech) occurs when the
stimulator touches the languages area. As the neurosurgeon stimulates
different areas of the brain and receives responses, it is common for
him to place numbered markers on the cortex highlighting regions to
avoid. When our probe is attached to the stimulator, we can obtain
the position of the tip during stimulations and immediately produce a
color-coded visualization highlighting these important areas.
![]()
![]() In most cases, one of the technicians sketches on paper where the grid
is located on the cortex as a reference during the week of monitoring.
Using our navigational system, we touch each grid point with the
Flashpoint probe and obtain the positions in model coordinates. Below
is the rendered image with the grid points in red. The doctors
monitoring the grid responses have reported that our images were very
helpful in drawing a correspondence between grid numbers and positions
on the cortex.
In most cases, one of the technicians sketches on paper where the grid
is located on the cortex as a reference during the week of monitoring.
Using our navigational system, we touch each grid point with the
Flashpoint probe and obtain the positions in model coordinates. Below
is the rendered image with the grid points in red. The doctors
monitoring the grid responses have reported that our images were very
helpful in drawing a correspondence between grid numbers and positions
on the cortex.
The electrodes can also be used to directly stimulate the surface of the cortex to map out the position of the motor and sensory corticies. In one case, we created a visualization where we colored the grid points depending on whether they were adjacent to motor cortex, sensory cortex, or the seizure focus. The neurosurgeon reported that the color-coding was very useful as he moved our probe over the cortex when planning out the region to resect.
Selected publications of the project
![]() W.E.L. Grimson,
G.J. Ettinger,
T. Kapur,
M.E. Leventon,
W.M. Wells III,
R. Kikinis.
"Utilizing Segmented MRI Data in Image-Guided Surgery."
In IJPRAI, 1996.
[color postscript 13.0M]
W.E.L. Grimson,
G.J. Ettinger,
T. Kapur,
M.E. Leventon,
W.M. Wells III,
R. Kikinis.
"Utilizing Segmented MRI Data in Image-Guided Surgery."
In IJPRAI, 1996.
[color postscript 13.0M]
![]() W.E.L. Grimson,
T. Lozano-Perez,
W.M. Wells III,
G.J. Ettinger,
S.J. White,
and
R. Kikinis,
"An Automatic Registration Method for Frameless Stereotaxy,
Image Guided Surgery, and Enhanced Reality Visualization"
In Transactions on Medical Imaging, 1996.
[gzipped postscript 3.2M]
W.E.L. Grimson,
T. Lozano-Perez,
W.M. Wells III,
G.J. Ettinger,
S.J. White,
and
R. Kikinis,
"An Automatic Registration Method for Frameless Stereotaxy,
Image Guided Surgery, and Enhanced Reality Visualization"
In Transactions on Medical Imaging, 1996.
[gzipped postscript 3.2M]
Back to the MIT AI Lab page.